Tools & Software
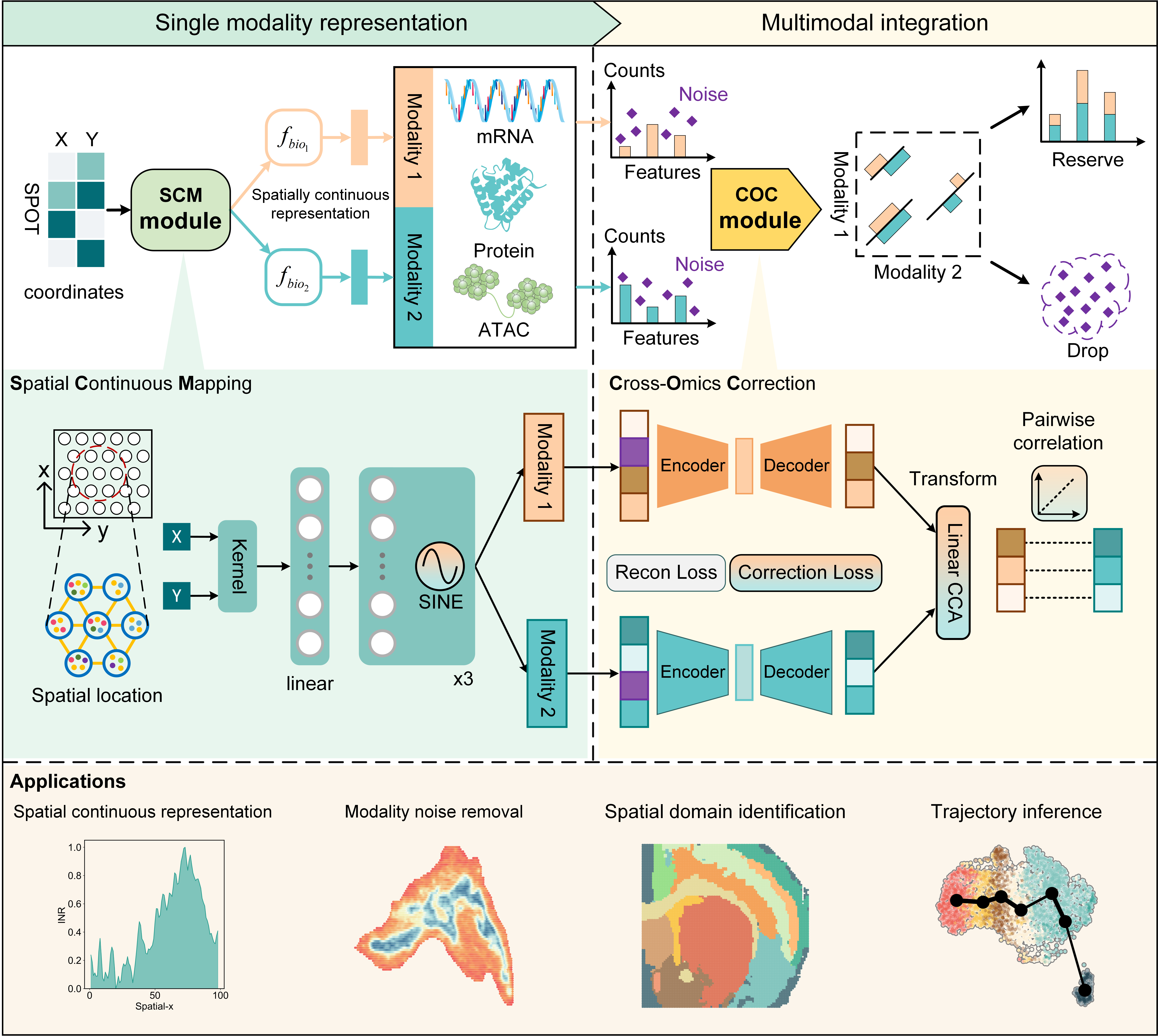
SpatialCOC
Spatial Multi-omics Integration Framework
SpatialCOC is a deep learning framework that deciphers cellular spatial organization and cross-omics relationships in spatial multi-omics data. It treats different omics modalities as continuous functions defined by their shared spatial coordinates, enabling it to uncover nonlinear correlations among these modalities. SpatialCOC excels in identifying region-specific continuous spatial domains and maintains batch-consistency across trajectory inferences.
Python Deep Learning Multi-omics Spatial Biology
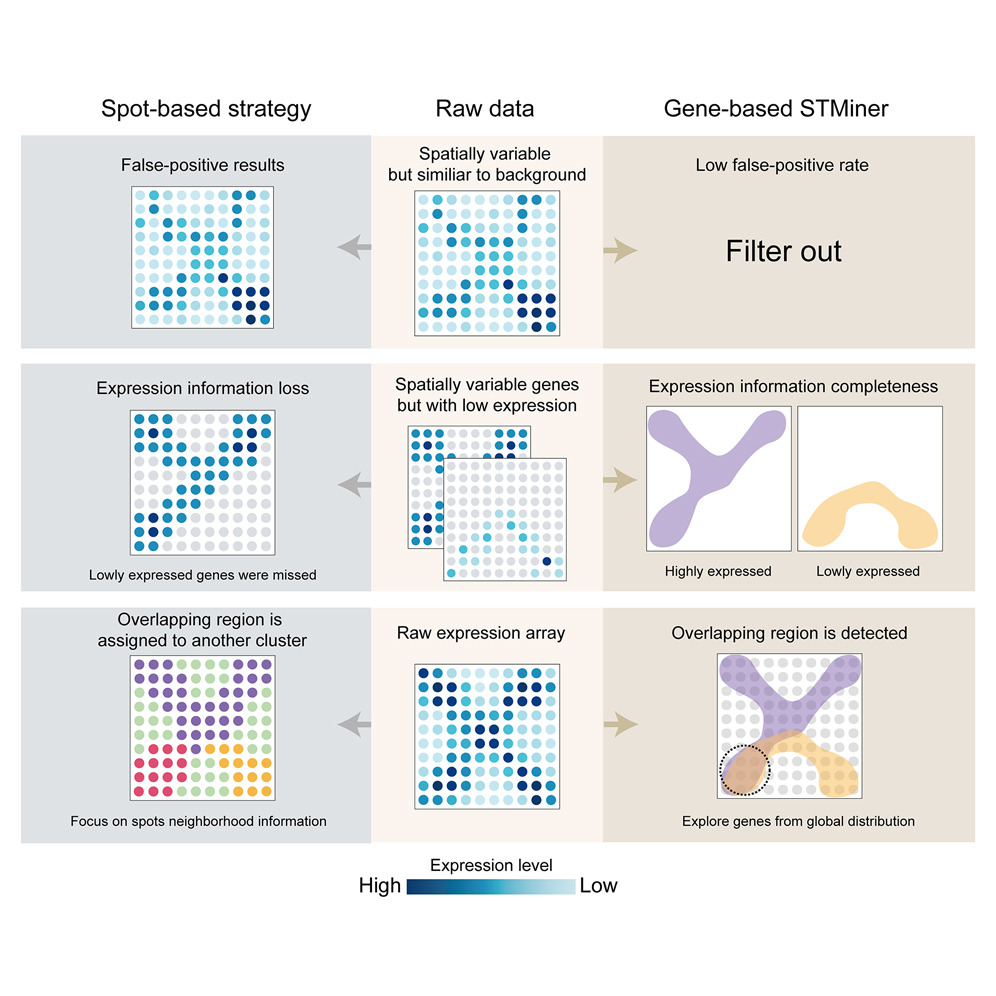
STMiner
Gene-centric Spatial Transcriptomics Analysis Tool
STMiner is a Python package for analyzing spatial transcriptomics data. It leverages 2D Gaussian mixture models and optimal transport theory to directly characterize the spatial distribution of genes, effectively mitigating the impacts of background bias and data sparsity. STMiner reveals key gene sets and spatial structures overlooked by spot-based analytic tools, facilitating novel biological discoveries in tumor tissues and complex spatial data.
Python Spatial Transcriptomics Bioinformatics